| Our group’s research is in the area of analytical mass spectrometry (MS). The major objectives of the research are to develop advanced methods and strategies, primarily based on MS, for the detection and characterization of biological molecules, including peptides, proteins, and metabolites, and to apply these methods to study real-world biological systems. The latter is being accomplished through active collaboration with researchers at the forefront of their disciplines.Our group is particularly interested in the analysis of biosystems, such as organelles, cells, tissues, and body fluids, by an interdisciplinary research approach that uses advanced analytical technologies and sophisticated bioinformatics tools to produce, interrelate, and integrate a large body of chemical and biological information on a biological system. Unlike medicine and systems biology, where more emphasis is on an overall understanding of the functions in a biological system, biosystems analysis focuses on the process of data generation and integration. Thus it can be considered as an important and fundamental part of systems biology and medicine.
Biosystems analysis requires analytical technologies having high sensitivity, specificity, and throughput to identify and quantify the chemical constituents of biosystems. Of the many available technologies, mass spectrometry is playing an increasingly important role in detection, identification and characterization of biological molecules. One major focus of our research is to develop enabling mass spectrometric tools for comprehensive proteome (the set of proteins that is or can be expressed by an organism) and metabolome (the set of all metabolites found in an organism, cell, or tissue) analysis.
Biosystems analysis is important in many areas of bioscience and biomedical research. For example, information on changes to the genome (the set of all genes in an organism), proteome, and metabolome of cells grown under different conditions (e.g., normal cell vs. diseased cell) can help us to understand better the biological functions of individual cellular components, which, in turn, can assist us to search for drug targets and drug candidates for better treatment and management of a disease. In the area of biomarker discovery, large-scale and systematic profiling of proteins and metabolites in biological samples, such as blood gathered from healthy and diseased populations, could result in the discovery of a number of chemicals that could potentially be used for early diagnosis and prognosis of a disease such as cancer.
Our current research activities include the following major projects:
- Development of analytical methods based on matrix-assisted laser desorption ionization (MALDI) and electrospray ionization (ESI) MS for proteome and metabolome analysis. The goals of this work are: 1) to generate accurate and comprehensive proteome and metabolome maps; 2) rapid quantification of proteome and metabolome changes; and 3) to carry out in-depth analysis of a sub-proteome, such as characterization of protein posttranslational modifications, or a sub-metabolome, such as all metabolites involved in a biological pathway.
- Development of nanoscale biological macromolecule manipulation and preparation methods for MS. The aim of this work is to improve mass spectrometric detection sensitivity and specificity for characterizing low abundance proteins and metabolites. We are particularly interested in analyzing very small amounts of samples, such as a single cell or primary cells procured from a tissue.
- Development and applications of proteomic and metabolomic approaches in disease biology study and biomarker discovery (e.g., cancer biology, cancer biomarker discovery).
|
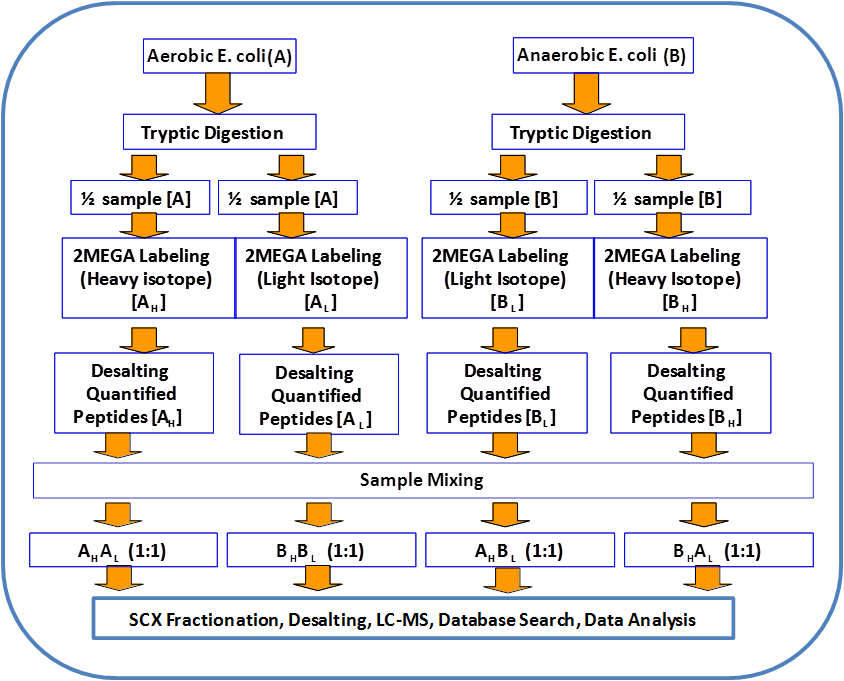 Andy Lo, et al., Anal. Chim. Acta. 2013, 795, 25. Andy Lo, et al., Anal. Chim. Acta. 2013, 795, 25.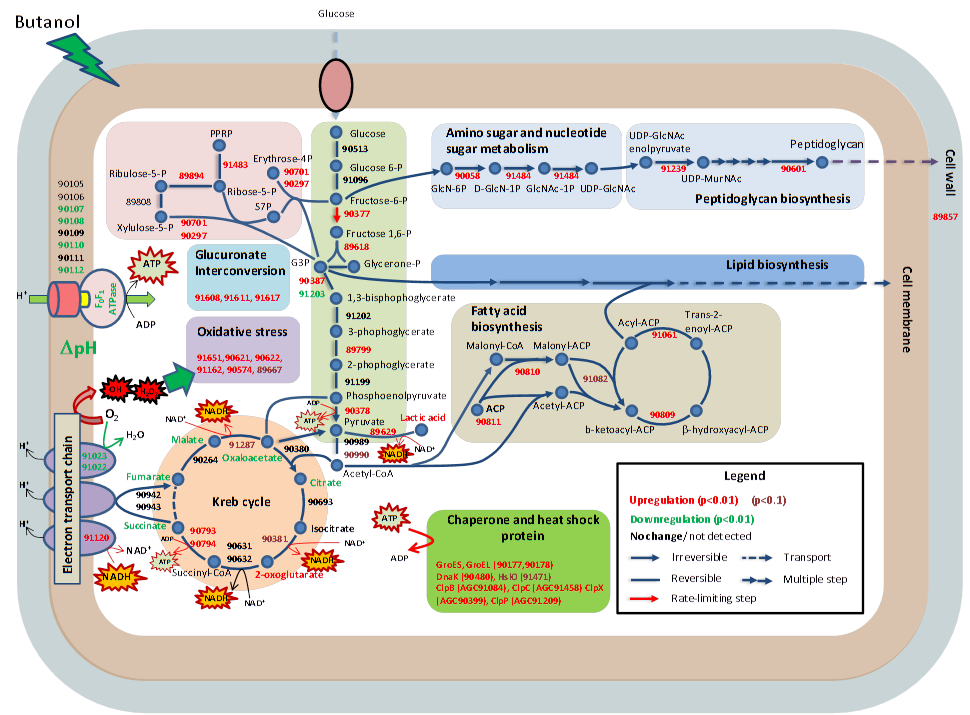
Feifei Fu, et al., J. Proteome Res. 2013, 12, 4478.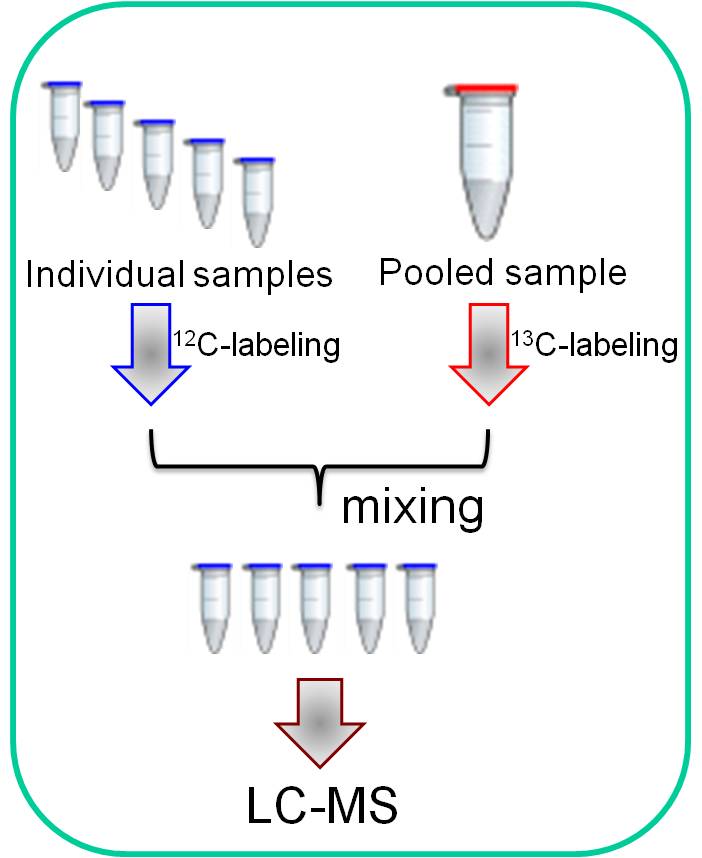 High-performance isotope labeling LC-MS metabolomics platform High-performance isotope labeling LC-MS metabolomics platform |

